To better understand its regulatory role, we characterized the targeted R-loops for each R-loop regulator by taking advantage of existing functional genomics data. The differential R-loop peaks following knockdown are likely targets of the corresponding regulator. In addition, R-loops may be directly modulated through chromatin or RNA binding activity of their regulators. Accordingly, we cataloged putative targeted R-loops for 52 validated R-loop regulators in human cells based on knockdown, ChIP-seq or CLIP-seq data.
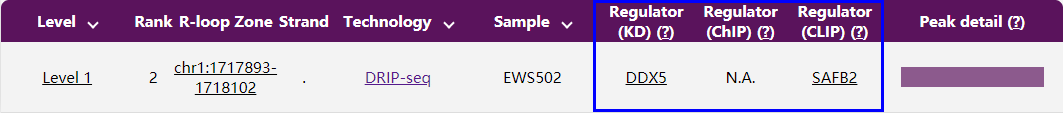
To get to know whether an R-loop region is targeted by a specific regulator, locate the R-loop by following the instructions detailed in Q6, all possible regulators will be listed in the last 2-4 columns, i.e., Regulator (KD), Regulator (ChIP), Regulator (CLIP).
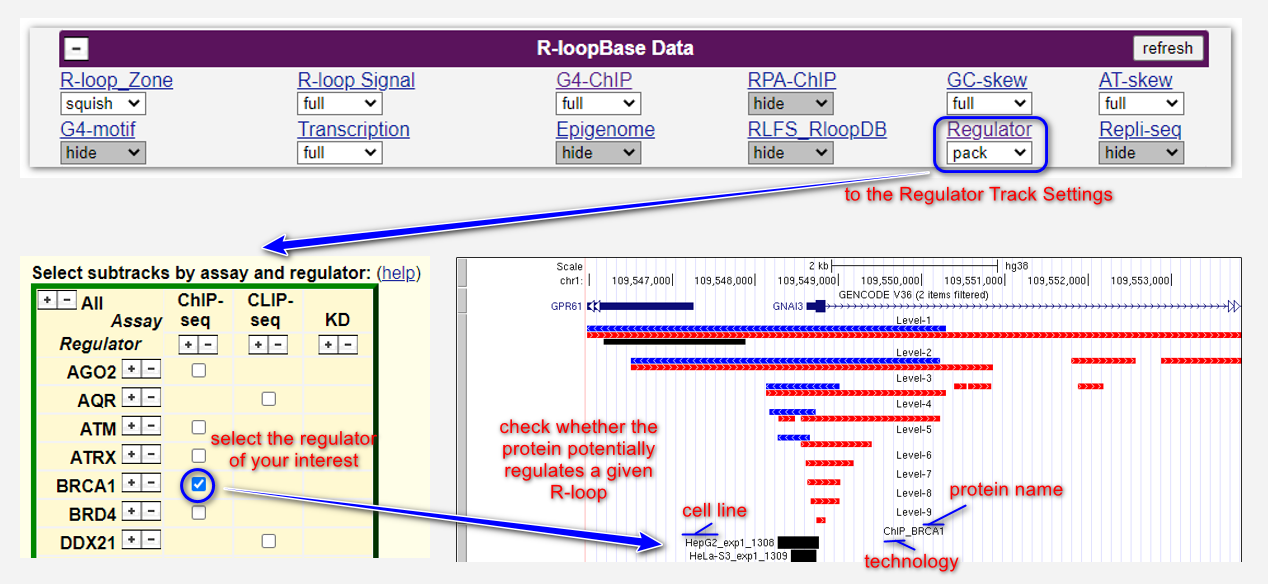
Alternatively, locate the R-loop region in genome browser. Open the Regulator Track settings to select specific genomics data for regulators of your interest, e.g., BRCA1 ChIP-seq data, by checking the corresponding box (see below). Whether an R-loop region is targeted by a specific regulator will be clearly shown on genome browser (see below).