You may achieve this via either of the following two methods.
(1) Type gene symbol (e.g., BRCA2) or hg38 genomic coordinates (e.g., chr1:1-100000) into the search box and then click "search" button. Here, only gene symbol and genomic coordinate are supported.

A summary report will return on top, showing number of R-loop zones detected by indicated technologies within the genomic locus of your interest.

At the bottom of the page, genomic coordinates of all R-loop zones are listed in a table, together with the confidence level, how and in which sample they were detected, and by which proteins they are potentially regulated. By default, level 1 R-loop zones are shown. You may access R-loop zones of other levels from the drop-down menu. You will be directed to Browser or Regulator page for more details by URLs linked to the genomic coordinates and regulators respectively.
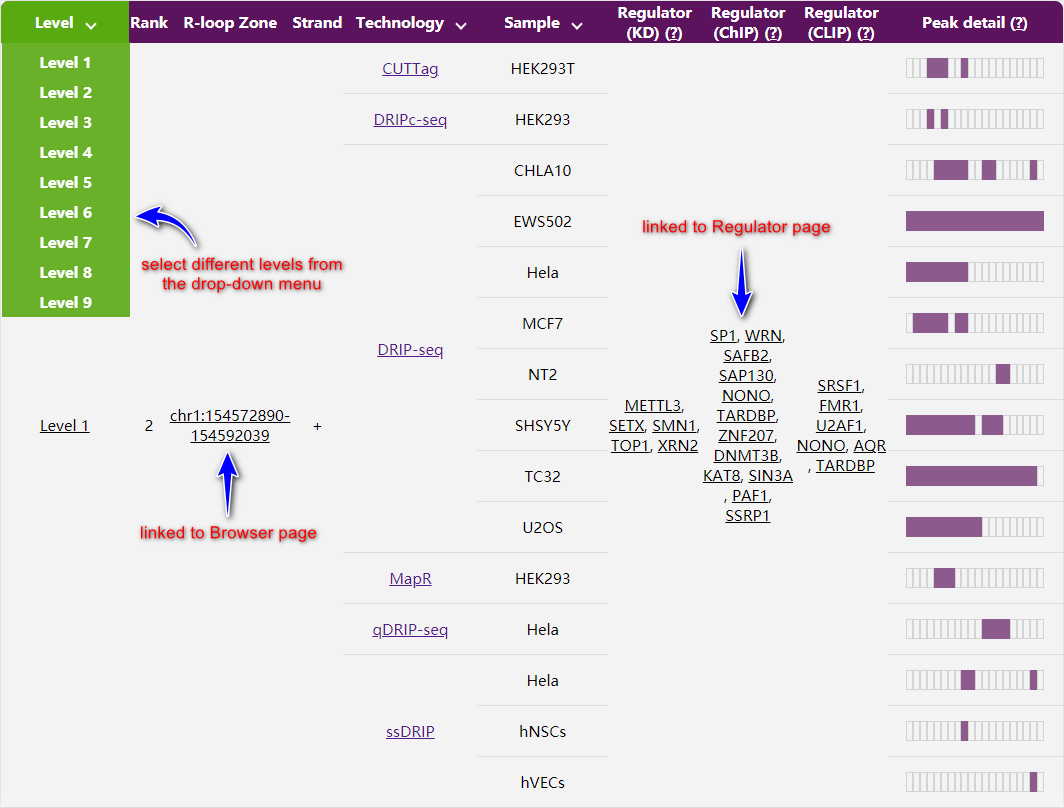
(2) Visit Browser from the navigation panel.

Type gene symbol (e.g., BRCA2), genomic coordinates or other IDs supported by UCSC genome browser (e.g., ENSEMBL, RefSeq and so on)

Visualize R-loop zones of different levels as well as the R-loop signals by configuring "R-loop Zone" and "R-loop Signal" tracks.