Q3. How have human R-loops been mapped on a genome-wide scale?
3.1. S9.6-based technologies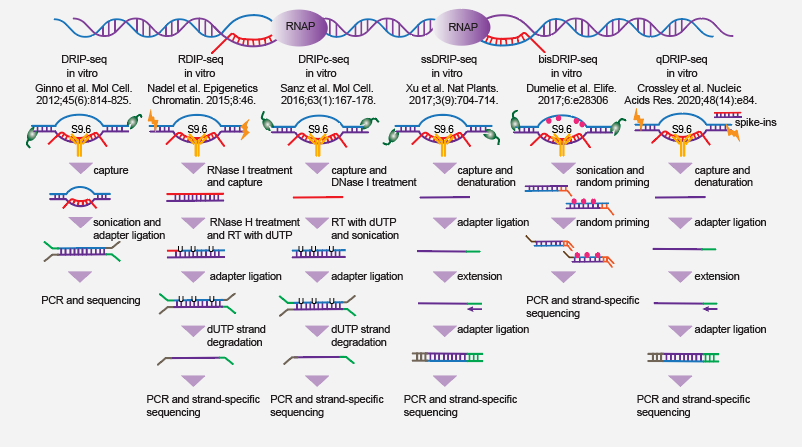
3.2. RNase H-based technologies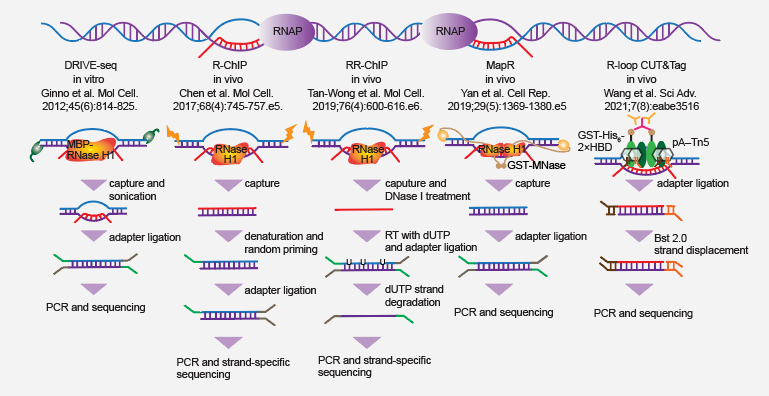
The vital biological functions of R-loops have fueled the development of two classes of genome-wide R-loop mapping technologies. The first takes advantage of the specificity of anti-RNA:DNA hybrid monoclonal antibody S9.6, while the second leverages (the RNA:DNA hybrid binding domain of) RNase H1 to capture R-loops.
3.1. S9.6-based technologies

- DRIP-seq, the first method for genome-wide mapping of R-loops, was initially developed by Fredric Chedin group in 2012 (PMID: 22387027). Basically, isolated genomic DNA is fragmented via a combination of restriction enzymes. R-loop-containing fragments are then enriched with S9.6 antibody and subjected to sonication, dsDNA end repair, adaptor ligation and PCR amplification for deep sequencing.
- RDIP-seq, another variant of DRIP-seq-like technology (PMID: 26579211), uses RNase I to remove any single-strand RNA from the sample, and sonication to minimize sequence bias by restriction digestion. Furthermore, template DNA is used for RNA-primed extension with dUTP and strand-specific library construction and sequencing.
- DRIPc-seq, as a modified version of DRIP-seq, was developed by Fredric Chedin group in 2016 (PMID: 27373332). Instead of sequencing dsDNA, the RNA moiety of R-loop structures is sequenced in a strand-specific manner.
- ssDRIP-seq, developed by Qianwen Sun group in 2017 (PMID: 28848233), uses more freqent cutters for DNA fragmentation. With further sonication, the resulting fragments after S9.6 enrichment presumably contain only the RNA:DNA hybrid but not the non-template strand. After denaturing, the template DNA is subjected to 3' ssDNA adaptor ligation, extension, 5' adaptor ligation, PCR and sequencing.
- bisDRIP-seq takes advantage of bisulfite for converting cytosines (Cs) into uracils (Us) within single-stranded DNA regions, and S9.6 for enriching R-loop structures. With subsequent sequencing and analyses, the direction and location of R-loop could be inferred from the distribution of C-to-U mutations. This method was developed by Samie Jaffrey group in 2017 (PMID: 29072160).
- qDRIP-seq, similar to ssDRIP, sequences the template DNA in strand-specific manner. However, the isolated DNA is fragmented with sonication instead of enzymatic digestion, and spike-ins might be introduced in for quantitatively measurement of R-loop formation. This method was reported by Karlene Cimprich group in 2020 (PMID: 32544226).
3.2. RNase H-based technologies

- DRIVE-seq was also developed by Fredric Chedin group at the same time of DRIP-seq (PMID: 22387027). The key difference with DRIP-seq is that human RNase H1 mutant instead of S9.6 is adopted for in vitro enrichment of RNA:DNA hybrids.
- R-ChIP, developed by Xiang-Dong Fu group, basically follows the typical ChIP-seq protocol (PMID: 29104020). Two key differences are introduced, i.e., catalytically-inactive human RNase H1 is exogenously expressed in cells for capturing R-loops in vivo, and the strand-specific sequencing library is prepared with random primer extension on template DNA.
- RR-ChIP, a derivative version of R-ChIP reported by Nick Proudfoot group (PMID: 31679819), sequences the RNA moiety instead of template DNA using dUTP strand-specific sequencing strategy.
- MapR, following the basic principle of CUT&RUN, utilizes RNase H to guide micrococcal nuclease to R-loop regions for cleavage, release and sequencing. This method was first reported by Kavitha Sarma group (PMID: 31665646), which avoids the step for generating stable cell lines expressing mutant RNase H as is the case for R-ChIP.
- R-loop CUT&Tag, based on the principle of CUT&Tag, utilizes hybrid binding domain of RNase H1 to guide Tn5 transposase for adaptor loading, followed by library construction and sequencing. This new method was recently developed by Kaiwei Liang group (PMID: 33597247)